Author
Author  Correspondence author
Correspondence author
Plant Gene and Trait, 2011, Vol. 2, No. 2 doi: 10.5376/pgt.2011.02.0002
Received: 24 Oct., 2011 Accepted: 25 Dec., 2011 Published: 16 Jan., 2012
Xiao et al., 2011, Cloning and Bioinformatics Analysis of Dihydroflavonol 4-Reductase Gene from Pigmented Potato, Plant Gene and Trait, Vol.2, No.2 7-14 (doi: 10.5376/pgt.2011.02.0002)
The main pigment which affects flower colors is the anthocyanin, and dihydroflavonol 4-reductase (DFR) is a key enzyme in anthocyanin synthesis. In this study, we employed the native pigmented potato varieties ‘Jianchuanhong’ and ‘Zhuanxinwu’ from Yunnan Province, and the introduced potato variety ‘Heimeiren’ as the experimental materials. The complete cDNAs sequence of DFR was cloned from their tubers epidermis by RT-PCR, and the bioinformatics analysis was carried out. The sequence analysis displayed that DFR gene of the three tested pigmented potato varieties encoded 381 amino acid and had a complete open reading frame (ORF). It also showed that DFR had a highly conserved NADP(H)-binding site and substrate specificity site. Homologous alignment result demonstrated that the nucleotide similarity and amino acid homology of the three kinds of pigmente potato species are 99.24%, 98.78%, respectively. And the amino acid homology of these pigmented potatoes with other Solanaceae plants, such as Nicotiana alata and Petunia x hybrida, were more than 86%. None DFR of these pigmente potatoes had signal peptide, obvious transmembrane structure region, we speculated that DFR might be a kind of hydrophilic protein which probably located in cytoplasm. We still found that α-helix and random coil were primary secondary structural components of DFR which belong to NADB-Rossmann superfamily.
The common varieties of potato (Solanum tuberosum L.) are yellow and white tuber, apart from them, there are also red, orange, purple, blue, black skin and other kinds of potato which is called Colored potato or Pigmented potato. Pigmented potato enrich anthocyanin, it has many beneficial effects on health, such as antioxidation, free radical scavenging, prevention of cardiovascular disease, reduction of blood lipid, anti-carcinogenesis (Eichhorn and Winterhalter, 2005; Lachman and Hamouz , 2005; Reddivari et al., 2007). The colors of pigmented potato tuber are mainly desided by two compounds carotenoids and anthocyanins. Carotenoids can cause the color of tuber skin and medullar tissues showing white or yellow (Brown, 2005). The accumulation of anthocyanins can make the color of the same tissues presenting red, blue, purple and so on.
The synthesis pathways of anthocyanins have been clearly understood, which was synthesized via Phenylalanine and methylmalonyl-CoA synthesis pathways (Tanaka et al., 1998). The accumulation of anthocyanins in flower and fruit of plants are regulated by some key enzymes and regulatory genes (Guo, 2011). Dihydroflavonol 4-reductase (DFR),a short chain reductase of DFR super family, is one of the key enzyme genes during the anthocyanin synthesis (Li et al., 2009; Martens et al., 2002 ). DFR could catalyze Dihydroflavonol (DHK), quercetol (DHQ) and myricetin becoming three sub-pathways and generated anthocyanins of orange pelargonidin, pink anthocyanin and purple delphinidin, respectively (Holton and Cornish, 1995).
In different species, based on the different substrates selectivity of DFR, which gave rise to the anthocyanins with different colors, and then gave distinct colors to all kinds of species (Liu et al., 2005). For example, DFR is able to restore three kinds of substrate efficiently in gerbera (Johnson et al., 2001), however, in petunidin and orchid, DFR can efficiently restore DHQ and DHM, while DHK can’t be restored. Therefore, orange pelargonidin can’t be generated in petunidin and orchid (Forkmann and Ruhnau, 1987; Johnson et al., 1999); whereas, the DFR of red potato can restore DHK (De et al., 2003).In different varieties, the temporal and spatial expression patterns of DFR are controlled by many structural genes and regulatory genes, and they are always expressed coordinately with CHS and CHI (Wang and Peng, 2000).
DFR is encoded by a single gene, which has been cloned from maize, snapdragon, China aster, Arabidopsis and petunidin (Oreilly et al., 1985; Wu et al., 2002). The zone type of restriction fragment length polymorphism (RFLP) indicated that DFR was a single copy gene in both potato and tomato (De et al., 2003; Bongue-Bartelsman et al., 1994). In grape, crystal structure of DFR protein is a complex with three subunits and has a specific NADPH domain (Petit and Granier, 2007).
In this research, we employed the native pigmented potato varieties ‘Jianchuanhong’ and ‘Zhuanxinwu’ from Yunnan Province, and the introduced potato variety ‘Heimeiren’ as the experimental materials to clone the complete cDNAs sequence of DFR and carried out the bioinformatics analysis. The cloning of DFR gene would play a foundation for studying the anthocyanins biosynthetic pathway and the regulatory mechanisms and provided theories for new variety breeding of pigmented potato.
1 Results and Analysis
1.1 cloning the dfr gene of pigmented potato
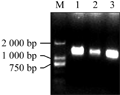
Took the first cDNA chain by reverse transcription reaction as the template, we amplified three pigmented potato varieties with degenerate primer. The result showed the size was almost the same (Figure 1). And then, the positive clones were selected by gel extraction purification, connection, transformation and colony PCR. The positive clones were extracted and sequenced.
|
|
1.2 The amino acid sequence analysis of DFR in pigmented potato
The result of DFR amino acid sequence blast showed that the NADPH binding site and the the specificity of substrate binding the amino sequence of Jianchuanhong, Zhuanxinwu and Heimeiren with DFR were all highly conserved (Figure 2). In the region of DFR combinding with substrates specifically, the amino acids site of 145 and 156 of three pigmented potato are D and E, respectivele, therefore, we predicted that it was corresponding to other species’ site of 134 and 145.
|
|
1.3 Physicochemical properties of DFR amino acid sequence in different species
The analysis of physicochemical properties of DFR gene by ProtParam showed in table 1. The results displayed that the number of amino acid residues, molecular weight and physicochemical property in Jianchuanhong, Zhuanxinwu and Heimeiren were almost the same. They wre identical with the data of potato cultivated species, wild species, the tobacco in the same family, the arabidopsis in same class from GenBank, while were different from wheat of monocotyledonous plant. The maximum content of amino acid among these amino acid sequences were Lys, Ala, Leu and Glu. None of them contains Pyl and Sec. According to the theory of protein stabilization, the unstable index of stable protein is lower than 40 (Zhang, 2009). It is hypothesized that DFR cDNA encoding amino acid sequence is a stable protein.
|
|
1.4 The similarities of DFR cDNA sequence and its amino acid sequence
The multiple alignments of DFR cDNA sequence and amino acid sequence showed in table 2. The results showed that there was high similarity in DFR cDNA sequence and the amino acid sequence in different plants. The average similarity of cDNA sequence in three tested pigmented potato varieties was 99.24%. Homology of the amino residues was 98.78%. At nucleic acid and protein level, the highest similarity is Jianchuanhong and potato cultivated species, which reached 100%. In the next place, the similarity among wild species, tobacco and petunia hybrida, from the same family, is above 86% (Table 2). In conclusion, DFR is very conservative among Solanaceae.
|
|
The phylogenetic tree was contructed by DNAman (Figure 3). The results demmonstrated that monocotyledon and dicotyledon formed two branches, the potato, tobacco and petunia hybrida were getting together. The homology of DFR in pigmented potato Jianchuanhong, Zhuanxinwu and potato cultivated species was highest. And that was also highest between“Heimeiren”and potato wild variety. All of them were in the same group to tobacco and petunia, they had a closest relationship, other dicotyledon ranked secondly, and monocotyledon, such as wheat, ranked thirdly.
|
|
1.5 Characteristics of DFR in hydrophilicity, hydrophobicity, trans-membrane domain and subcellular localization
SignalP analysis results showed that the DFR protein of the three tested pigmented potato varieties didn’t have signal peptide, which indicated that ir was not a secretory protein. ProtScale on-line analysis of both TMHMM and ExPASy indicated that DFR protein didn’t have obvious trans-membrane domain.
Prediction of hydrophilicity and hydrophobicity is a necessary procedure on predicting protein secondary structure and division of functional domain. ProtScale analysis results showed that the hydrophilicity /hydrophobicity of DFR protein in three tested pigmented potato varieties were 2.411 in maximum, -2.633 in minimum. The trend pictures of hydrophilicity /hydrophobicity were basically identical. The hydrophilicity /hydrophobicity analysis results of Jianchuanhong showed in Figure 4. According to the theory that the score of amino acid is lower, the hydrophilicity is stronger; the score of amino asid higher, the hydrophobicity is stronger. The number of hydrophilic amino acid residues is more than hydrophobicity amino acid residues in one peptide chain. Therefore, we could hypothesize that the DFR protein in the three tested pigmented potato varieties were hydrophilic proteins.
|
|
The prediction of subcellular localization of tested pigmented potato varieties with PSORT II showed that DFR of Jianchuanhong and Zhuanxinwu was likely to be localized in cytoplasm with a probability of 39.1%, and that of Heimeiren, the highest probability was 69.6%.
Protein is synthesized in ribosome, and this synthesis starts from cytoplasmic matrix. However, some proteins, such as secretory protein, transmembrane protein and glycoprotein, would synthesize the signal peptide consisting of several amino acids. Generally, the signal peptide is hydrophobic. Therefore, it can infiltrate to the lipid bilayer of endoplasmic reticulum. Because of the signal peptide, the ribosome can adhere to endoplasmic reticulum to synthesize protein. In the present study, DFR protein of these three tested pigmented potato varieties did not have this signal peptide and obvious trans-membrane domain. It might be a hydrophilic protein instead of a secretory protein. Moreover, it was localized in cytoplasm. All of these information indicated that DFR protein of potato was synthesized in cytoplasm.
1.6 The prediction of secondary structure and function domain of DFR
The prediction results of secondary structure of DFR by SOPMA showed that the secondary structure of DFR in the three tested pigmented potato varieties consisted of random coil, alpha helix, extended strand and beta turn. The results of prediction of secondary structure in Jianchuanhong were shown in Figure 5. The proportion of random coil, alpha helix, extended strand and beta turn were 43.19%, 36.91%, 12.83% and 7.07%, respectively. The proportion of secondary structure in Zhuanxinwu, Heimeiren and Jianchuanhong is basically identical. The secondary structure proportion of random coil and alpha helix was above 80% in the three tested pigmented potato varieties, which further verified that DFR was a stable stabilizing protein.
Protein domain is the structural basis of protein performing functions. The prediction results of protein domain of DFR by Pfam25.0 showed that it had only one domain in DFR protein from 20 to 269 in amino acid region (Figure 5). The 175th amino acid residue is the active site. Therefore, we could hypothesize that the 175th amino acid residues was the functional domain of DFR protein. The result of DFR belonged to NADB-Rossmann superfamily was confirmed by CDD on-line Prediction on NCBI (Figure 6).
|
|
|
|
2 Discussion
DFR gene was cloned from tuber epidermis of three potato varieties with different color, including red Jianchuanhong, purple Zhuanxinwu and black Heimeiren. The sequence of ribonucleotide and amino, NADH binding sites and sites binding with substrate specifically were highly conserved, which indicated that there were other key enzyme genes including regulatory gene and environmental factors which control the difference of tint in pigmented potato as well as DFR gene. Homology of cDNA and amino sequence between Yunnan local variety Jianchuanhong and Zhuanxinwu and potato cultivated species was 100% and 99%, respectively. So they might be different ecotypes with ordinary potato cultivated species consanguinity, and might be developed in specific climate and ecological environment of Yunnan. The homology between Heimeiren from America and wild species was highest in different species. We hypothesized that Heimeiren might has consanguinity of wild species. The DFR gene of Heimeiren was from the same group with tobacco and petunia and had the closest relationship. Other dicotyledon ranked secondly, and monocotyledon such as wheat ranked thirdly. The results were in agreement with classification in plant morphology, which was reasonable to speculate the systemic evolution of species with the data of bioinformatics.
In various species, DFR amino acid sequence has high homology. It is highly homologous with the binding site of NADPH (VTGAAGFIGSWLVMRLLERGY) and the amino acid sequence combining with substrate specifically (TLDVQEDQKLFYDETSWSDLDFIYAK) (Polashock et al., 2002). The 134th and 145th amino acid in this area affected substrate specificity of DFR directly. The 132th amino acid in majority of species is D (Aspartic acid) or N (Asparagine). Except petunia, the glutamic acid E in 145th amino acid of all dicotyledon with the substrate DHK was highly conserved and this was not the case of petunia. For petunia, the 134th amino acid sequence was D. DFR can not produce pelargonidin with DHK as the substrate, while most plants with N at 134th amino acid can produce pelargonidin (Johnson et al., 2001). The corresponding site of these three pigmented potatoes cloned in this study was D, and the results of prophase study suggested that they can produce pelargonidin (De et al., 2003). Petunia and potato belonged to Solanaceae, and the corresponding site was D. However, petunia was not able to generate pelargonidin, while potato could do that. Then, a series of enzymes in metabolic pathway of various species and the mechanisms involved in interaction of intermediate product need to be further investigated.
The research of Cheng (Cheng et al., 2010) and his group indicated that DFR of petunia is localized in mitochondria. It was a transmembrane protein with a signal peptide and transit peptides. However, DFR of three pigmented potato varieties were localized in without signal peptide and obvious trans-membrane domain. So DFR had different characteristics compared with different class in Solanaceae. The further investigation will be needed in this field.
3 Materials and Methods
3.1 Materials and reagents
Jianchuanhong and Zhuanxinwu samples were from Yunnan Province. Jianchuanhong was red skin and white medullar with red circles. Zhuanxinwu was purple skin and yellow medullar with purple vascular loop. Heimeiren was introduced variety from America, with black skin and medullar. All the three tested pigmented potato varieties were cultivated on the farm of Yunnan Agricultural University in the same cultivation conditions.
In multiple sequence alignment, the sequence of nucleotide and corresponding amino acid of DFR were from database of nucleonic acid and protein on NCBI.
The reagents includeing RNA extraction reagents TransZol Up and two step reverse transcription kit were purchased from TransGen Biotech (Beijing). DNA glue recovery kit and LB medium were purchased from Sangon Biotech (Shanghai). DNA Marker DL-2000, pMD18-T vector and JM109 competent cell were purchased from TaKaRa (Dalian).
3.2 RNA isolation and the cloning of DFR
Referring to the instruction book of TransZol Up RNA extraction reagent and two-step method reverse transcription kit, total RNA was extracted from epidermis of these three pigmented potato, and then the first strand cDNA was synthesized by reverse transcription. The primer of DFR is DFR (F): 5’-CTGAAAATGGCAAGTGAAG-3’ and DFR (R): 5’-GGTTCTAGATTTCACCATTGG -3’. RT-PCR reaction system: first strand cDNA 2 μL; DFR(F) 1 μL; DFR(R) 1 μL; 2xTransTaqTM High Fidelity (HiFi) PCR SuperMix 25 μL; ddH2O was added up to 50 μL. The amplification program was devised as follows: initial denaturation for 4 min at 94℃, denaturing at 94℃ for 1 min, primers annealing at 50.5℃ for 45 s, extension at 72℃ for 70 s, then final extension was at 72℃ for 12 min after 35 cycles. A target fragment was recovered by using agarose DNA purification kit, and then 4 μL recovery products and 1 μL pMD-18T vector were ligated at 16℃ for 4 h. Ligation product was added into 50μL JM109 competent cells to transform, and positive recombinants, which were sent to Sangon Biotech (Shanghai) Co., Ltd. to bi-directional sequencing, were detected by colony PCR. It was spliced by DNAMAN, acquiring full length of cDNA of DFR gene.
3.3 Bioinformatics analysis
DFR cDNA sequence and the composition and physicochemical properties of its’ amino acid sequence were analyzed online by ProtParam (http://web.expasy.org/protparam/) in the toolkit of ExPASy. ORF was found using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) online tools in NCBI. Sequence similarity searches were completed by Blastn and Blastp in NCBI (http://blast.ncbi.nlm.nih.gov/). Homology multiple blasting of DFR nucleotide and amino acid sequence were finished by DNAMAN, and phylogenetic tree was constructed according to the result. Signal peptide of protein was analyzed online using SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/). Trans-membrane domain was analyzed online by TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and ProtScale (http://web.expasy.
org/protscale/) in the toolkit of ExPASy. Hydrophily and hydrophobicity of DFR protein were predicted online by ProtScale (http://web.expasy.org/protscale/) in the toolkit of ExPASy. Bubcellular localization was investigated using PSORT II Prediction (http://psort.hgc.jp/form2.html). Functional domain of DFR protein was predicted online using Pfam25.0 (http://pfam.sanger.ac.uk/) and CDD in NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Authors’ contributions
JPX and HCG were responsible for experimental design and the experiment direction; JPX carried out most of the work; QW and WLL took part in the work; JPX and HCG were responsible for data analysis, paper writing and modification. All of them had read the final version of this paper and agreed with the authors’ credits.
Acknowledgement
This study was supported by Chinese National Modern Agricultural Technology System (CARS-10) and Yunnan Province Scientific and Technological Project (2009BB010) together.
Reference
Brown C.R., 2005, Antioxidants in potato, American Journal of Potato Research, 82(2): 163-172
http://dx.doi.org/10.1007/BF02853654
Bongue-Bartelsman M., O’Neill S.D., Tong Y., and Yoder J. I., 1994, Characterization of the gene encoding dihydroflavonol 4-reductase in tomato, Gene, 138(1-2): 153-157
http://dx.doi.org/10.1016/0378-1119(94)90799-4
Chen D.Z., Zhou J.Y., and Li P., 2010, Bioinformatics analysis of dihydroflavonol 4-reductase, Shengwu Jishu Tongbao (Biotechnology Bulletin), 12: 206-212
De J. W.S., De J. D.M., De J. H., Kalazich J., and Bodis M., 2003, An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.), Theor Appl Genet, 107(8): 1375-1383
http://dx.doi.org/10.1007/s00122-003-1395-9
Eichhorn S., and Winterhalter., 2005, Anthocyanins from pigmented potato (Solanum tuberosum L.) varieties., Food Research International, 38(Issues 8-9): 943-948
Forkmann G., and Ruhnau B., 1987, Distinct substrate specificity of dihydroflavonol 4-reductase from flowers of petunia hybrida, Z. Naturforsch, 42c: 1146-1148
Guo J.Y., Li Y.P., Fu Y.F., Huang C.T., Zhou W., and Gao F., 2011, Correlations among the enzyme activity and gene expression of dihydroflavonol 4-reductase and anthocyanin accumulation in purple-fleshed sweet potato, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 44(8): 1736-1744
Holton T.A., and Cornish E.C., 1995, Genetics and biochemistry of anthocyanin biosynthesis, Plant Cell 7(7): 1071-1083
http://dx.doi.org/10.1105/tpc.7.7.1071 http://dx.doi.org/10.2307/3870058
Johnson E.T., Yi H., Shin B., Oh B.J., Cheong H., and Choi G., 1999, Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type Anthocyanins, Plant J., 19(1): 81-85 http://dx.doi.org/10.1046/j.1365-313X.1999.00502.x
Johnson E.T., Ryu S., Yi H., Shin B., Cheong H., and Choi G., 2001, Alteration of a single amino-acid changes the substrate specificity of dihydroflavonol 4-reductase, Plant J., 25(3): 325-333
http://dx.doi.org/10.1046/j.1365-313x.2001.00962.x
Liu J., Feng Q.F., and Zhang J., 2005, Dihydroflavonol 4-reductase Gene (DFR) and flower color modulation, Zhiwu Shenglixue Tongxun (Plant Physiology Journal), 41(6): 715-719
Lachman J., and Hamouz K., 2005, Red and purple coloured potatoes as a significant antioxidant source in human nutrition – a review, Plant Soil Environ, 51(11): 477-482
Li C.L., Cui G.X., Xu Z.R., and Li Y.H., 2009, Research advances on dihydroflavonol 4 -reductase gene, Shengwu Jishu Tongxun (Letters in Biotechnology), 20(3): 442-445
Martens S., Teeri T., and Forkmann G., 2002, Heterologous expression of dihydroflavonol 4-reductases from various plants, FEBS letters, 531(3): 453-458
http://dx.doi.org/10.1016/S0014-5793(02)03583-4
Oreilly C., ShePherd N.S., Pereira A., Schwarz-Sommer Z., Bertram I., Robertson D.S., Peterson P.A., and Saedler H., 1985, Molecular cloning of the a1 locus of zea mays using the transposable elements En and Mu1, EMBOJ, 4(4): 877-882
Reddivari L., Vanamala J., Chintharlapalli S., Safe S.H., and Creighton Miller Jr. J., 2007, Anthocyanin fraction from potato extracts is cytotoxic to prostate cancer cells through activation of caspase-dependent and caspase-independent pathways, Carcinogenesis, 28(10): 2227-2235
http://dx.doi.org/10.1093/carcin/bgm117
Polashock J.J., Griesbaeh I.U., Sullivan R.F., and Voma N., 2002, Cloning of a cDNA encoding the cranberry dihydroflavonol-4-reductase(DFR) and expression in transgenic tobacco, Phat Science, 163(2): 241-251
Petit P., Granier T., Langlois B., d’Estaintot., Manigand C., Bathany K., Schmitter J.M., Lauvergeat V., Hamdi S., and Gallois B., 2007, Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis, Journal of Molecular Biology, 368(5): 1345-1357
http://dx.doi.org/10.1016/j.jmb.2007.02.088
Tanaka Y., Tsuda S., and Kusumi T., 1998, Metabolic engineering to modify flower color, Plant and Cell Physiology, 39(11): 1119-1126
Wang Z.K., and Peng Z.H., 2000, Advances in genetic engineering of ornamental plant, Forest Research, 13(1): 97-102
Wu S.H., and Zhang D.S., 2002, The research advances on dfr gene of anthocyanin biosynthesis, Fujian Linxueyuan Xuebao (Journal of Fujian College of Forestry), 22(2): 189-192
Zhang Y.L., Zhang Z.J., Yang F.S., Mahesh K., Yuan H., and Luo S.P., 2009, Cloning and bioinformatics analysis of PmNHX1 gene from Xinjiang Halophyte Plantago maritima, Zhongguo Shengwu Gongcheng Zazhi (China Biotechnology), 29(1): 27-33
. PDF(541KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jiping Xiao
. Qiong Wang
. Wanlin Li
. Huachun Guo
Related articles
. Pigmented potato
. Dihydroflavonol 4-reductase
. cDNA cloning
. Sequences analysis
Tools
. Email to a friend
. Post a comment